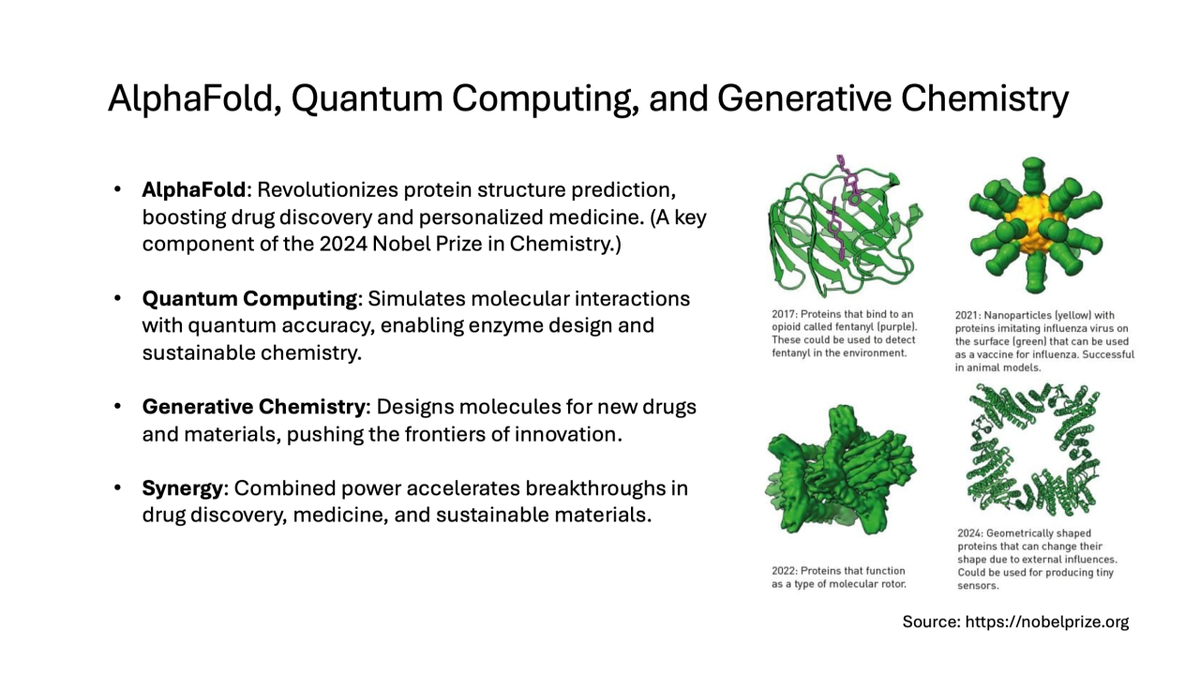
AlphaFold and 2024 Nobel Prize in Chemistry
In 2024, Demis Hassabis and John Jumper were awarded the Nobel Prize in Chemistry for their work with AlphaFold. This recognition reflects the significance of AlphaFold’s contribution to protein science and drug discovery, as it now helps scientists worldwide understand and manipulate proteins more effectively. AlphaFold’s success shows how AI, initially developed for games, can be applied to solve some of the most challenging problems in science, underscoring the potential of AI to drive innovation across fields.
AlphaGo: In 2016, DeepMind's AlphaGo made headlines by defeating the world champion in the game of Go, a highly complex board game with more possible moves than atoms in the observable universe. AlphaGo used deep neural networks and reinforcement learning, demonstrating AI’s ability to master complex strategic thinking beyond human capability.
AlphaZero: Building on AlphaGo, AlphaZero was developed as a more generalized AI model. Unlike AlphaGo, which was trained specifically on Go, AlphaZero taught itself to play games like Go, chess, and shogi without prior human knowledge. This model used reinforcement learning to rapidly learn strategies, displaying a more flexible and powerful approach to AI.
AlphaFold: DeepMind’s greatest breakthrough in science came with AlphaFold, an AI system designed to predict protein structures—an essential problem in biology that had eluded researchers for decades. Understanding protein structures is crucial because it allows scientists to understand biological functions and design new drugs. AlphaFold achieved remarkable accuracy in predicting protein shapes based on amino acid sequences, solving the "protein-folding problem" and making a transformative impact on medicine, biochemistry, and biotechnology.
AlphaFold’s Strengths and Limitations
Strengths: AlphaFold, developed by DeepMind, uses advanced deep learning algorithms to predict protein structures with high accuracy based on amino acid sequences. It has been instrumental in unlocking insights into protein folding, which is crucial for understanding biological functions, drug design, and disease mechanisms.
Limitations: However, AlphaFold relies on classical computing and statistical methods, which approximate molecular interactions. This makes it challenging to account fully for the complex quantum mechanical interactions that govern molecular behavior, especially when it comes to accurately predicting binding affinities, reaction dynamics, or subtle changes in protein conformations under different conditions.
The Role of Quantum Computing in Molecular Simulation
Quantum Precision: Quantum computers have the potential to simulate molecular interactions at a level of precision that classical computers struggle to achieve. By leveraging qubits, quantum computers can represent complex quantum states and model the behavior of electrons and atoms in ways that are more true to quantum mechanics.
Enhanced Drug Discovery: With quantum computing, it would be possible to simulate drug-protein interactions, enzyme dynamics, and reaction pathways more accurately, leading to better predictions of drug efficacy and potential side effects. This precision could complement AlphaFold’s predictions by providing finer details of how proteins behave and interact with other molecules in a quantum context.
Integrating AlphaFold with Quantum Computing
Hybrid Classical-Quantum Models: A promising approach would be to use AlphaFold’s classical machine learning model to predict initial protein structures and then refine these predictions using quantum simulations for specific tasks, like binding affinity calculations or protein-ligand interactions. This hybrid model could combine AlphaFold’s structural predictions with the quantum-level detail offered by quantum computers.
Quantum-Enhanced Machine Learning: Quantum computing could also enhance machine learning models by optimizing algorithms and processing large amounts of data in parallel. Future versions of AlphaFold could use quantum algorithms to analyze complex biological data and explore protein structure spaces that are computationally inaccessible with classical methods alone.
Generative Chemistry
What is Generative Chemistry? Generative chemistry involves using AI and machine learning models to design new molecules with desired properties, such as higher efficacy, lower toxicity, or improved stability. Generative models, like those used in generative adversarial networks (GANs) or reinforcement learning, can "generate" novel molecules and test them virtually for specific applications.
Quantum Computing in Generative Chemistry: Quantum computing can bring generative chemistry to a new level by accurately simulating molecular interactions, which is essential for predicting the behavior of novel molecules. This capability is crucial for tasks like identifying new drug candidates or creating advanced materials. Quantum computers could evaluate generated molecules with unprecedented accuracy, allowing scientists to screen compounds more efficiently and with higher confidence in the results.
AlphaFold + Quantum Generative Chemistry: Combining AlphaFold’s structural insights with quantum-powered generative chemistry could be a game-changer in drug design. For example, AlphaFold could predict the 3D structure of a target protein, and a quantum-augmented generative model could design molecules specifically tailored to interact with that structure. Quantum simulations would then validate these molecules’ interactions at an atomic level, predicting binding affinities, toxicity, and overall drug efficacy.
The Synergy of AI and Quantum Computing in Biomedicine
Accelerating Drug Discovery: This combination of AlphaFold, quantum computing, and generative chemistry could make the drug discovery process faster and more precise, allowing researchers to screen billions of compounds virtually before any lab testing. It could significantly reduce the time and cost associated with bringing new drugs to market.
Designing Custom Proteins and Enzymes: Beyond drug discovery, quantum-enhanced generative models could be used to design custom proteins or enzymes with unique properties, useful in fields such as synthetic biology, bioengineering, and environmental science.
Exploring Uncharted Chemical Spaces: The synergy between AI and quantum computing could allow scientists to explore vast, uncharted chemical spaces. Generative chemistry, powered by quantum simulations, could lead to the discovery of entirely new classes of compounds that are optimized for specific functions, such as ultra-efficient catalysts or biodegradable materials.
Summary
AlphaFold, quantum computing, and generative chemistry represent three pillars of next-generation computational science:
- AlphaFold provides structural predictions of proteins.
- Quantum Computing offers precise simulations of molecular interactions, bringing quantum-level accuracy to biochemistry.
- Generative Chemistry creates novel molecules with desired properties, essential for drug design and material science.
Together, these technologies could create a powerful pipeline for scientific discovery, leveraging AlphaFold’s structural predictions, quantum computing’s accuracy, and generative models' creativity to transform industries from biomedicine to materials science. This marriage of AI and quantum computing, especially in fields like protein folding and molecular design, is poised to unlock new scientific frontiers and make previously impossible discoveries feasible.


Member discussion: